Contents
py4DSTEM Parallax Masks Notebook
Imports¶
%matplotlib widget
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
import py4DSTEM
import ipywidgets
from IPython.display import display
style = {'description_width': 'initial'}Load Data¶
file_path = 'data/'
file_data_01 = file_path + 'parallax_apoferritin_simulation_100eA2_01.h5'
file_data_02 = file_path + 'parallax_apoferritin_simulation_100eA2_02.h5'
dataset_01 = py4DSTEM.read(file_data_01)
dataset_02 = py4DSTEM.read(file_data_02)
dataset = py4DSTEM.DataCube(np.hstack((dataset_01.data,dataset_02.data)),calibration=dataset_01.calibration)
datasetDataCube( A 4-dimensional array of shape (24, 48, 128, 128) called 'datacube',
with dimensions:
Rx = [0.0,10.666666666666666,21.333333333333332,...] A
Ry = [0.0,10.666666666666666,21.333333333333332,...] A
Qx = [0.0,0.1538085070134974,0.3076170140269948,...] mrad
Qy = [0.0,0.1538085070134974,0.3076170140269948,...] mrad
)Basic Visualizations¶
mean_diffraction_pattern = dataset.get_dp_mean().data
mean_diffraction_pattern /= mean_diffraction_pattern.max()
bright_field_mask = mean_diffraction_pattern > 0.5
bright_field_mask_indices_vertical, bright_field_mask_indices_horizontal = np.where(bright_field_mask)
reciprocal_sampling = dataset.calibration.get_Q_pixel_size()
reciprocal_units = dataset.calibration.get_Q_pixel_units()
reciprocal_kwargs = {"pixelsize":reciprocal_sampling,"pixelunits":reciprocal_units,"scalebar":{"length":5},"ticks":False}
virtual_bright_field_images = dataset[:,:,bright_field_mask].transpose(2,0,1)
gridscan_sampling = dataset.calibration.get_R_pixel_size()
real_kwargs = {"pixelsize":gridscan_sampling/10,"pixelunits":"nm","scalebar":{"color":"black","length":10},"ticks":False}def return_axial_image_and_mask(axial_r):
""" """
axial_bf_indices = np.unravel_index(
np.argwhere(
np.linalg.norm(
np.argwhere(np.ones_like(bright_field_mask)) - np.array(bright_field_mask.shape)[None]/2,
axis=1
) < axial_r
),
bright_field_mask.shape
)
axial_bf_image = dataset[:,:,axial_bf_indices[0],axial_bf_indices[1]].sum((-2,-1))
bright_field_mask_rgb = np.zeros(bright_field_mask.shape+(3,))
# cyan
bright_field_mask_rgb[bright_field_mask,1:] =1
# magenta
bright_field_mask_rgb[axial_bf_indices[0],axial_bf_indices[1],0]=1
bright_field_mask_rgb[axial_bf_indices[0],axial_bf_indices[1],1]=0
bright_field_mask_rgb[axial_bf_indices[0],axial_bf_indices[1],2]=1
return axial_bf_image, bright_field_mask_rgb
def update_axial_image_and_mask(change):
axial_r = change["new"]
axial_bf_image, bright_field_mask_rgb = return_axial_image_and_mask(axial_r)
axial_artists[0].set_data(bright_field_mask_rgb)
_axial_bf_image, _vmin, _vmax = py4DSTEM.visualize.return_scaled_histogram_ordering(axial_bf_image)
axial_artists[1].set_data(_axial_bf_image)
axial_artists[1].set_clim(vmin=_vmin, vmax=_vmax)
fig_axial.canvas.draw_idle()
return None
axial_r_slider = ipywidgets.IntSlider(min=1, max=26, step=1, style=style, description="axial virtual BF radius")
axial_r_slider.observe(update_axial_image_and_mask,names='value')with plt.ioff():
dpi = 72
fig_axial = plt.figure(figsize=(675/dpi, 360/dpi), dpi=dpi)
gs = GridSpec(2, 2, wspace=0.05)
ax_bf_disk = fig_axial.add_subplot(gs[:, 0])
ax_axial = fig_axial.add_subplot(gs[0, 1])
ax_incoherent = fig_axial.add_subplot(gs[1, 1])
axial_bf_image, bright_field_mask_rgb = return_axial_image_and_mask(axial_r=1)
py4DSTEM.show(
bright_field_mask_rgb,
figax=(fig_axial,ax_bf_disk),
**reciprocal_kwargs,
title="bright field disk mask",
)
py4DSTEM.show(
axial_bf_image,
figax=(fig_axial,ax_axial),
**real_kwargs,
cmap='Greys',
title="virtual (axial) bright field image",
bordercolor="magenta",
borderwidth=2,
)
py4DSTEM.show(
virtual_bright_field_images.sum(0),
figax=(fig_axial,ax_incoherent),
**real_kwargs,
cmap='Greys',
title="incoherent bright field image",
bordercolor="cyan",
borderwidth=2,
)
gs.tight_layout(fig_axial)
axial_artists = [ax_bf_disk.get_images()[0],ax_axial.get_images()[0]]
fig_axial.canvas.resizable = False
fig_axial.canvas.header_visible = False
fig_axial.canvas.footer_visible = False
fig_axial.canvas.toolbar_visible = True
fig_axial.canvas.layout.width = '675px'
fig_axial.canvas.layout.height = '390px'
fig_axial.canvas.toolbar_position = 'bottom'display(
ipywidgets.VBox(
[
axial_r_slider,
fig_axial.canvas
],
layout=ipywidgets.Layout(align_items='center',width='675px'))
)VBox(children=(IntSlider(value=1, description='axial virtual BF radius', max=26, min=1, style=SliderStyle(desc…Iterative Alignment Bins¶
def return_bf_bins(bf_mask, bin_value, shift=0):
""" """
_xy_inds = np.argwhere(bf_mask)
_xy_center = (_xy_inds - np.mean(_xy_inds, axis=0)).astype("float")
_xy_inds_binned = np.round(_xy_center / bin_value + shift).astype("int")
_xy_vals_binned = np.unique(_xy_inds_binned, axis=0)
_inds_order = np.argsort(np.sum(_xy_vals_binned**2, axis=1))
masks = np.zeros((_inds_order.shape[0],)+bf_mask.shape)
for ind, bin_ind in enumerate(_inds_order):
sub = np.logical_and(
_xy_inds_binned[:, 0] == _xy_vals_binned[bin_ind, 0],
_xy_inds_binned[:, 1] == _xy_vals_binned[bin_ind, 1],
)
indexing_i, indexing_j = _xy_inds[sub].T
masks[ind,indexing_i,indexing_j]=1
return masks
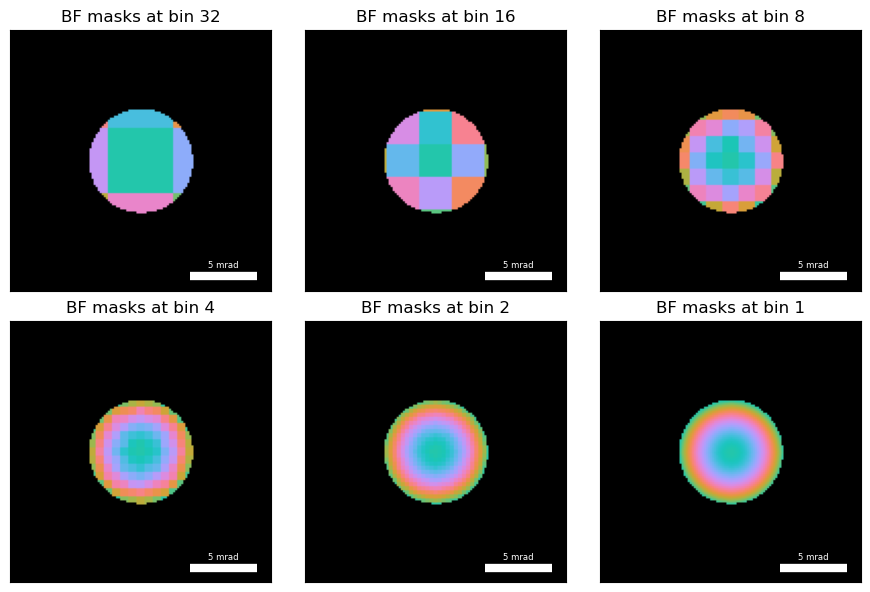
def return_bf_bins_rgb(bf_mask, bin_value, shift=0):
""" """
bins=return_bf_bins(bf_mask, bin_value, shift=shift)
n = bins.shape[0]
hue_angles = np.linspace(0.0, 2.0 * np.pi, n, endpoint=False)
complex_arr = np.tensordot(np.exp(1j*hue_angles),bins,axes=1)
return py4DSTEM.visualize.Complex2RGB(complex_arr)
bins_32 = return_bf_bins_rgb(bright_field_mask,32)
bins_16 = return_bf_bins_rgb(bright_field_mask,16)
bins_8 = return_bf_bins_rgb(bright_field_mask,8)
bins_4 = return_bf_bins_rgb(bright_field_mask,4)
bins_2 = return_bf_bins_rgb(bright_field_mask,2)
bins_1 = return_bf_bins_rgb(bright_field_mask,1)with plt.ioff():
py4DSTEM.show(
[
[bins_32, bins_16, bins_8],
[bins_4, bins_2, bins_1]
],
axsize=(3,3),
**reciprocal_kwargs,
title=[f"BF masks at bin {it}" for it in [32,16,8,4,2,1]]
)
fig_masks = plt.gcf()
fig_masks